Note
Go to the end to download the full example code
Basic Data Operations¶
A simple example showing how to download a dataset from neurovault and perform basic data operations. The bulk of the nltools toolbox is built around the Brain_Data() class. This class represents imaging data as a vectorized features by observations matrix. Each image is an observation and each voxel is a feature. The concept behind the class is to have a similar feel to a pandas dataframe, which means that it should feel intuitive to manipulate the data.
Download pain dataset from neurovault¶
Here we fetch the pain dataset used in Chang et al., 2015 from neurovault. In this dataset there are 28 subjects with 3 separate beta images reflecting varying intensities of thermal pain (i.e., high, medium, low). The data will be downloaded to ~/nilearn_data, and automatically loaded as a Brain_Data() instance. The image metadata will be stored in data.X.
from nltools.datasets import fetch_pain
data = fetch_pain()
Dataset created in /home/runner/nilearn_data/chang2015_pain
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_10_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_10_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_10_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_11_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_11_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_11_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_12_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_12_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_12_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_13_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_13_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_13_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_14_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_14_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_14_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_15_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_15_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_15_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_16_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_16_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_16_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_17_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_17_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_17_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_18_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_18_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_18_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_19_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_19_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_19_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_1_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_1_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_1_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_20_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_20_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_20_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_21_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_21_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_21_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_22_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_22_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_22_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_23_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_23_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_23_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_24_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_24_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_24_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_25_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_25_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_25_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_26_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_26_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_26_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_27_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_27_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_27_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_28_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_28_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_28_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_2_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_2_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_2_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_3_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_3_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_3_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_4_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_4_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_4_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_5_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_5_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_5_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_6_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_6_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_6_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_7_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_7_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_7_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_8_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_8_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_8_Medium.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_9_High.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_9_Low.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/504/Pain_Subject_9_Medium.nii.gz ...
...done. (0 seconds, 0 min)
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
Load files¶
Nifti images can be easily loaded simply by passing a string to a nifti file. Many images can be loaded together by passing a list of nifti files. For example, on linux or OSX systmes, the downloads from fetch_pain() will be stored in ~/nilearn_data. We will load subject 1’s data.
# NOTES: Need to figure out how to get path to data working on rtd server
# from nltools.data import Brain_Data
# import glob
#
# sub1 = Brain_Data(glob.glob('~/nilearn_data/chang2015_pain/Pain_Subject_1*.nii.gz'))
Basic Brain_Data() Operations¶
Here are a few quick basic data operations. Find number of images in Brain_Data() instance
print(len(data))
84
Find the dimensions of the data. images x voxels
print(data.shape())
(84, 238955)
We can use any type of indexing to slice the data such as integers, lists of integers, or boolean.
print(data[[1,6,2]])
nltools.data.brain_data.Brain_Data(data=(3, 238955), Y=(0, 0), X=(3, 57), mask=MNI152_T1_2mm_brain_mask.nii.gz)
Calculate the mean for every voxel over images
data.mean()
nltools.data.brain_data.Brain_Data(data=(238955,), Y=(0, 0), X=(0, 0), mask=MNI152_T1_2mm_brain_mask.nii.gz)
Calculate the standard deviation for every voxel over images
data.std()
nltools.data.brain_data.Brain_Data(data=(238955,), Y=(0, 0), X=(0, 0), mask=MNI152_T1_2mm_brain_mask.nii.gz)
Methods can be chained. Here we get the shape of the mean.
print(data.mean().shape())
(238955,)
Brain_Data instances can be added and subtracted
new = data[1]+data[2]
Brain_Data instances can be manipulated with basic arithmetic operations Here we add 10 to every voxel and scale by 2
data2 = (data+10)*2
Brain_Data instances can be copied
new = data.copy()
Brain_Data instances can be easily converted to nibabel instances, which store the data in a 3D/4D matrix. This is useful for interfacing with other python toolboxes such as nilearn
data.to_nifti()
<nibabel.nifti1.Nifti1Image object at 0x7f377be289a0>
Brain_Data instances can be concatenated using the append method
new = new.append(data[4])
Any Brain_Data object can be written out to a nifti file
data.write('Tmp_Data.nii.gz')
Images within a Brain_Data() instance are iterable. Here we use a list comprehension to calculate the overall mean across all voxels within an image.
[x.mean() for x in data]
[0.86061627, 0.5476546, 0.7457299, 0.25735068, -0.06096518, 0.08957473, -0.1356948, -0.5642196, -0.5352845, 0.23520215, 0.073372886, 0.011462674, 0.2969622, -0.28074002, -0.18138647, 0.08738991, 0.05743886, 0.019836975, -0.036790244, -0.1305875, -0.029930038, 0.105367, 0.02061516, 0.008520396, 0.3590906, 0.040400386, 0.2065466, 0.36504507, 0.029532516, -0.026943885, -0.10943584, -0.3174775, -0.21014933, 0.30044985, -0.056538206, 0.13816155, 0.21413556, -0.20611525, -0.08890725, 0.56508994, 0.24812311, 0.35720587, 0.15037484, 0.05586886, -0.061988655, 0.8838908, 0.33612254, 0.45791322, 0.13033451, -0.1309218, -0.007976429, 0.045336694, -0.038327135, -0.09149013, 0.113571614, -0.05455998, 0.007123048, 0.051181536, -0.027594494, 0.007650891, -0.061395787, -0.09653523, -0.06447013, -0.003625449, -0.027816378, -0.027183175, 0.020997265, -0.05787875, -0.07143228, 0.33068687, 0.077882566, 0.2516894, 0.19467102, 0.10458751, 0.12041345, 0.24733096, 0.057716135, 0.21319006, 0.28260896, -0.18303774, 0.07332882, -0.0004305779, -0.24120164, -0.1059538]
Basic Brain_Data() Plotting¶
There are multiple ways to plot data. First, Brain_Data() instances can be converted to a nibabel instance and plotted using any plot method such as nilearn.
from nilearn.plotting import plot_glass_brain
plot_glass_brain(data.mean().to_nifti())
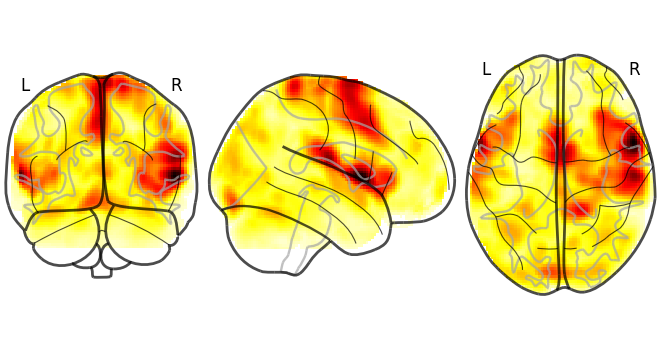
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f377b451f40>
There is also a fast montage plotting method. Here we plot the average image it will render a separate plot for each image. There is a ‘limit’ flag which allows you to specify the maximum number of images to display.
data.mean().plot()
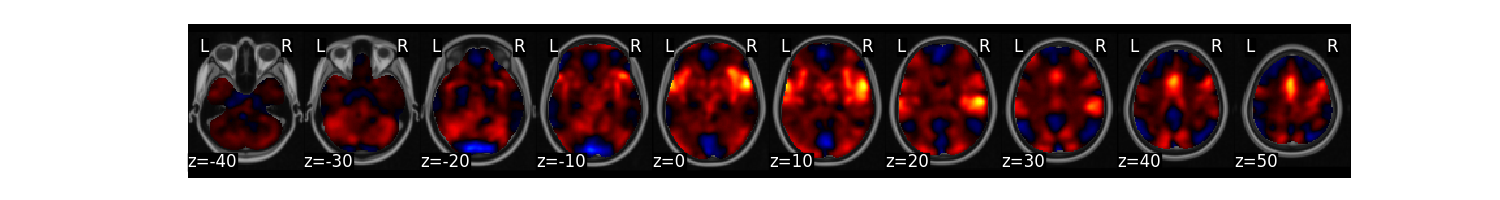
Total running time of the script: (1 minutes 11.902 seconds)