Note
Go to the end to download the full example code
Neurovault I/O¶
Data can be easily downloaded and uploaded to neurovault using pynv, a python wrapper for the neurovault api.
Download a Collection¶
Entire collections from neurovault can be downloaded along with the accompanying image metadata. You just need to know the collection ID. Data will be downloaded to the path specified in the ‘data_dir’ flag or ‘~/nilearn_data’ by default. These files can then be imported into nltools as a Brain_Data() instance.
from nltools.datasets import download_collection
from nltools.data import Brain_Data
metadata,files = download_collection(collection=2099)
mask = Brain_Data(files,X=metadata)
Dataset created in /home/runner/nilearn_data/2099
Downloading data from http://neurovault.org/media/images/2099/Neurosynth%20Parcellation_0.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/2099/Neurosynth%20Parcellation_1.nii.gz ...
...done. (0 seconds, 0 min)
Downloading data from http://neurovault.org/media/images/2099/Neurosynth%20Parcellation_2.nii.gz ...
...done. (0 seconds, 0 min)
Download a Single Image from the Web¶
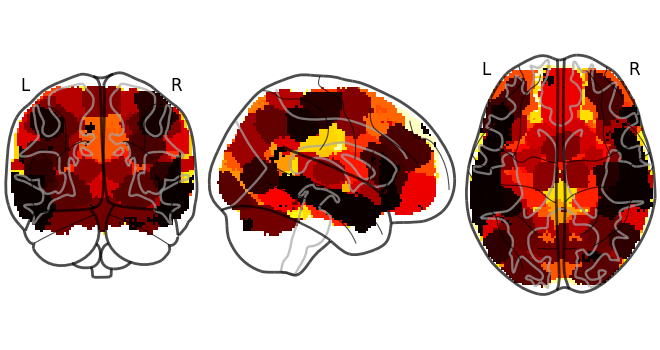
It’s possible to load a single image from a web URL using the Brain_Data load method. The files are downloaded to a temporary directory and will eventually be erased by your computer so be sure to write it out to a file if you would like to save it. Here we plot it using nilearn’s glass brain function.
from nilearn.plotting import plot_glass_brain
mask = Brain_Data('http://neurovault.org/media/images/2099/Neurosynth%20Parcellation_0.nii.gz')
plot_glass_brain(mask.to_nifti())
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f377bc94520>
Upload Data to Neurovault¶
There is a method to easily upload a Brain_Data() instance to neurovault. This requires using your api key, which can be found under your account settings. Anything stored in data.X will be uploaded as image metadata. The required fields include collection_name, the img_type, img_modality, and analysis_level. See https://github.com/neurolearn/pyneurovault_upload for additional information about the required fields. (Don’t forget to uncomment the line!)
api_key = 'your_neurovault_api_key'
# mask.upload_neurovault(access_token=api_key, collection_name='Neurosynth Parcellation',
# img_type='Pa', img_modality='Other',analysis_level='M')
Total running time of the script: (0 minutes 4.543 seconds)