Note
Go to the end to download the full example code
Decomposition¶
Here we demonstrate how to perform a decomposition of an imaging dataset. All you need to do is specify the algorithm. Currently, we have several different algorithms implemented from scikit-learn (‘PCA’,’ICA’,’Factor Analysis’,’Non-Negative Matrix Factorization’).
Load Data¶
First, let’s load the pain data for this example. We need to specify the training levels. We will grab the pain intensity variable from the data.X field.
from nltools.datasets import fetch_pain
data = fetch_pain()
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/nilearn/maskers/nifti_masker.py:108: UserWarning: imgs are being resampled to the mask_img resolution. This process is memory intensive. You might want to provide a target_affine that is equal to the affine of the imgs or resample the mask beforehand to save memory and computation time.
warnings.warn(
Center within subject¶
Next we will center the data. However, because we are combining three pain image intensities, we will perform centering separately for each participant.
data_center = data.empty()
for s in data.X['SubjectID'].unique():
sdat = data[data.X['SubjectID']==s]
data_center = data_center.append(sdat.standardize())
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:246: UserWarning: Numerical issues were encountered when centering the data and might not be solved. Dataset may contain too large values. You may need to prescale your features.
warnings.warn(
Decomposition with Factor Analysis¶
We can now decompose the data into a subset of factors. For this example, we will use factor analysis, but we can easily switch out the algorithm with either ‘pca’, ‘ica’, or ‘nnmf’. Decomposition can be performed over voxels or alternatively over images. Here we perform decomposition over images, which means that voxels are the observations and images are the features. Set axis=’voxels’ to decompose voxels treating images as observations.
n_components = 5
output = data_center.decompose(algorithm='fa', axis='images',
n_components=n_components)
Display the available data in the output dictionary. The output contains a Brain_Data instance with the brain factors (e.g., output[‘components’]), the feature by component weighting matrix (output[‘weights’]), and the scikit-learn decomposition object (output[‘decomposition_object’]. The Decomposition object contains the full set of information, including the parameters, the components, and the explained variance.
print(output.keys())
dict_keys(['decomposition_object', 'components', 'weights'])
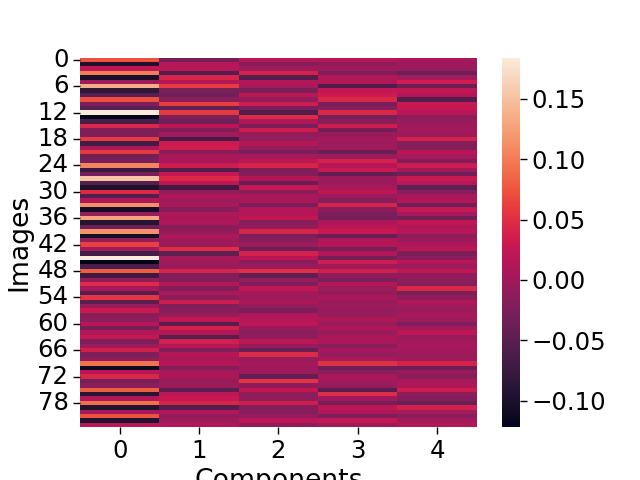
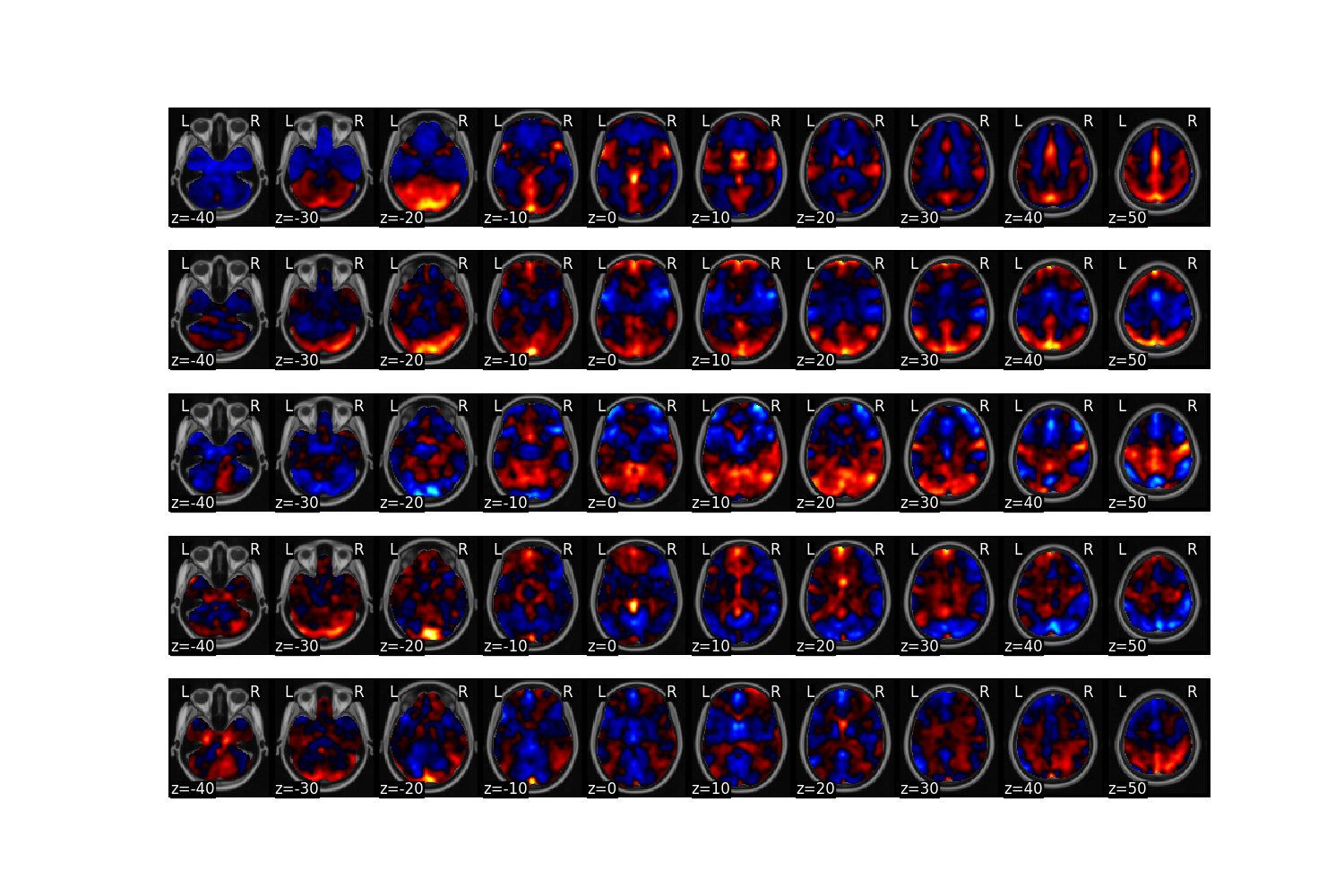
Next, we can plot the results. Here we plot a heatmap of how each brain image loads on each component. We also plot the degree to which each voxel loads on each component.
import seaborn as sns
import matplotlib.pylab as plt
with sns.plotting_context(context='paper', font_scale=2):
sns.heatmap(output['weights'])
plt.ylabel('Images')
plt.xlabel('Components')
output['components'].plot(limit=n_components)
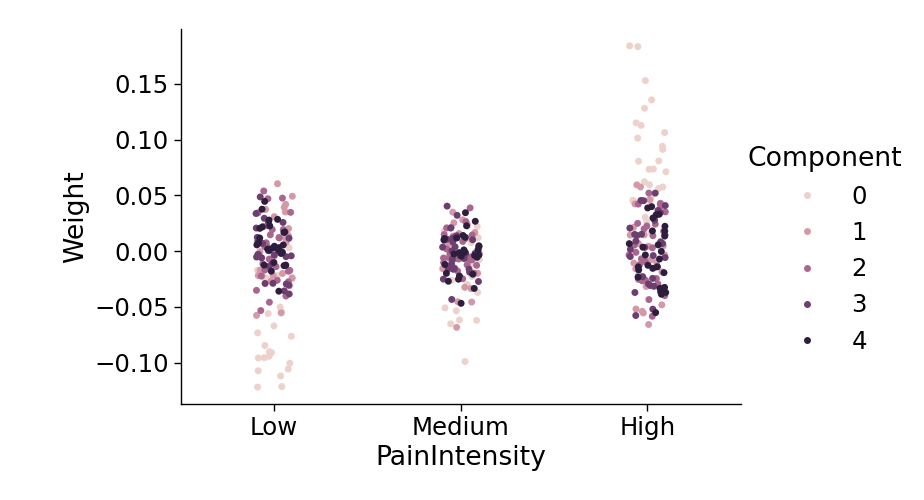
Finally, we can examine if any of the components track the intensity of pain. We plot the average loading of each component onto each pain intensity level. Interestingly, the first component with positive weights on the bilateral insula, s2, and ACC monotonically tracks the pain intensity level.
import pandas as pd
wt = pd.DataFrame(output['weights'])
wt['PainIntensity'] = data_center.X['PainLevel'].replace({1:'Low',
2:'Medium',
3:'High'}
).reset_index(drop=True)
wt_long = pd.melt(wt,
value_vars=range(n_components),
value_name='Weight',
var_name='Component',
id_vars='PainIntensity')
with sns.plotting_context(context='paper', font_scale=2):
sns.catplot(data=wt_long,
y='Weight',
x='PainIntensity',
hue='Component',
order=['Low','Medium','High'],
aspect=1.5)
Total running time of the script: (0 minutes 50.307 seconds)